Animal models play a critical role in the study of Huntington’s disease (HD), for example to elucidate underlying molecular mechanisms or to develop novel therapeutic venues. One of the first transgenic mouse models of HD, the R6/2 line, has been described in 1996 and has since become one of the most studied models of the disease (Mangiarini et al. 1996; 8898202 ![]() ; Li, Popovic & Brundin 2005; 16389308
; Li, Popovic & Brundin 2005; 16389308 ![]() ). R6/2 mice express the human exon 1 under control of the human huntingtin promoter at around 75% of the endogenous levels. The expression of the exon 1 with a CAG repeat length of around 115 evokes rapid disease progression and animals display multiple HD-associated neuropathological changes. First signs of disease can occur as early as three weeks and mice are severely impaired by the age of 8-12 weeks. The expected life span is 13-16 weeks (Li, Popovic & Brundin 2005; 16389308
). R6/2 mice express the human exon 1 under control of the human huntingtin promoter at around 75% of the endogenous levels. The expression of the exon 1 with a CAG repeat length of around 115 evokes rapid disease progression and animals display multiple HD-associated neuropathological changes. First signs of disease can occur as early as three weeks and mice are severely impaired by the age of 8-12 weeks. The expected life span is 13-16 weeks (Li, Popovic & Brundin 2005; 16389308 ![]() ).
).
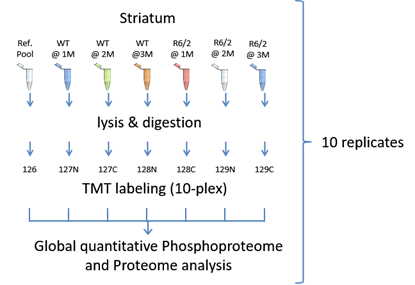
We were interested in the changes occurring in the proteomic landscape as well as in phosphoproteomic signaling networks in the R6/2 mouse model as compared to wild type mice. To this end, we performed comprehensive quantitative phosphoproteome and proteome analyses of striatum of one, two, and three months old mice of both genotypes. A total of nine or more samples per group was analyzed according to the experimental layout shown above. To allow for accurate quantification, peptides of each sample were labelled using tandem mass tags (TMT) and replicates of different conditions were combined. Each combined sample contained an aliquot of a pooled sample consisting of 12 sample lysates. This pooled sample was intended to serve as a reference to allow for quantitative comparison between all combined samples.
The results of this proteomics study have now been added to HD Proteome Base. The user-friendly web portal available within HDinHD allows the researcher to query for proteins and to visualize their expression. HD Proteome Base is based on MaxQB, which was developed in collaboration with the Max-Planck Institute of Biochemistry in Martinsried, Germany (Schaab et al., MCP 2012, M111. 014068, PMID: 22301388 ![]() ). Whereas MaxQB serves as generic repository for quantitative proteomics data, HD Proteome Base focusses on HD-related experiments. In addition to this study, HD Proteome Base also allows users query for proteins and to visualize their expression across the CAG repeat length series and across different tissues generated by the Mouse Allelic Series project (Langfelder et al., Nat Neurosci. 2016; 26900923
). Whereas MaxQB serves as generic repository for quantitative proteomics data, HD Proteome Base focusses on HD-related experiments. In addition to this study, HD Proteome Base also allows users query for proteins and to visualize their expression across the CAG repeat length series and across different tissues generated by the Mouse Allelic Series project (Langfelder et al., Nat Neurosci. 2016; 26900923 ![]() ). By augmenting HD Proteome Base with this new R6/2 study (raw data available for download from PRIDE as project PXD013771), we hope to help multiple research teams across the globe to gain a better understanding of the huntingtin biology and potentially contribute to the development of new therapies for HD.
). By augmenting HD Proteome Base with this new R6/2 study (raw data available for download from PRIDE as project PXD013771), we hope to help multiple research teams across the globe to gain a better understanding of the huntingtin biology and potentially contribute to the development of new therapies for HD.
Access the HD Proteome Base:


