In release v1.7, we leverage large-scale HD perturbation data to introduce a new type of gene set within HDSigDB, the HD and HD-related gene set enrichment library. Curation of experimental data from the HD perturbation literature captures the nature of the a) HD model; b) perturbation; and c) readout/result, each according to a well-characterized ontology. “Perturbation gene sets” are constructed by grouping a set of perturbed genes that share a readout – e.g. aggregation – and result – e.g. the treatment effect (perturbed HD vs HD) shows amelioration or exacerbation of a phenotype as compared to the genotype effect (HD vs WT).
To provide further granularity and interpretability, we breakdown each of these groups independently by 2 experimental factors related to the HD Model:
- Does the experiment utilize an in vitro, in vivo or ex vivo HD model?
- Does the HD Model contain a fragment or full-length mHTT construct?
As a result, using aggregation as an example readout, we have the following distinct gene sets created from HD perturbations for the aggregation readout:
| Gene Set Name | Number of Genes |
|---|---|
| Perturbations ameliorate the aggregation phenotype within Fragment mHtt HD models [HDinHD.org Perturbation Studies] | 543 |
| Perturbations exacerbate the aggregation phenotype within Fragment mHtt HD models [HDinHD.org Perturbation Studies] | 424 |
| Perturbations exacerbate the aggregation phenotype within Full-Length mHtt HD models [HDinHD.org Perturbation Studies] | 20 |
| Perturbations ameliorate the aggregation phenotype within Full-Length mHtt HD models [HDinHD.org Perturbation Studies] | 62 |
| Perturbations ameliorate the aggregation phenotype within in vitro HD models [HDinHD.org Perturbation Studies] | 423 |
| Perturbations exacerbate the aggregation phenotype within in vitro HD models [HDinHD.org Perturbation Studies] | 320 |
| Perturbations exacerbate the aggregation phenotype within in vivo HD models [HDinHD.org Perturbation Studies] | 220 |
| Perturbations ameliorate the aggregation phenotype within in vivo HD models [HDinHD.org Perturbation Studies] | 221 |
| Perturbations exacerbate the aggregation phenotype within ex vivo HD models [HDinHD.org Perturbation Studies] | 1 |
| Perturbations ameliorate the aggregation phenotype within ex vivo HD models [HDinHD.org Perturbation Studies] | 3 |
We have added a new facet to the Gene Set Enrichment Library portal called Gene Set Source that allows users to filter for different types of gene sets, including Perturbation gene sets.
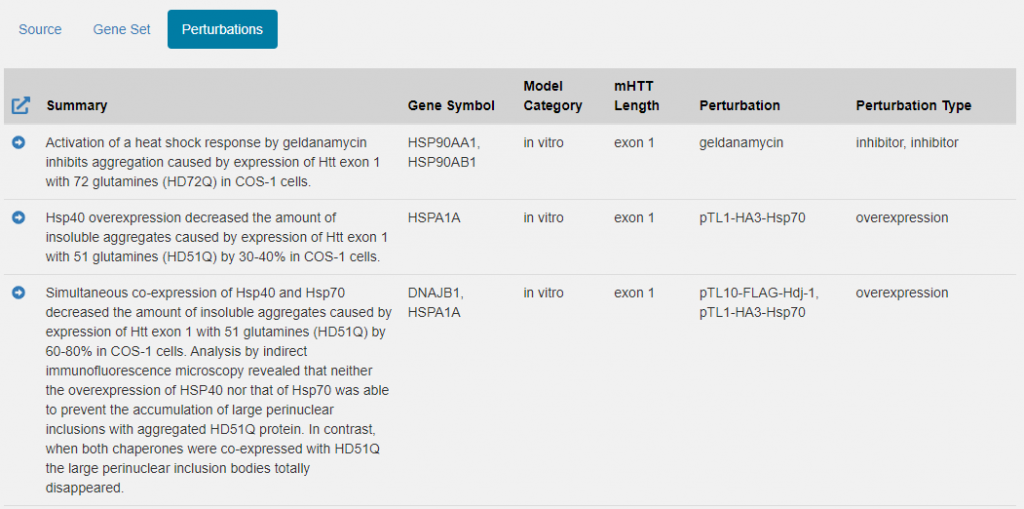
Provenance for most gene sets to date refer to a single source – either a publication or an omics study. Perturbations gene sets, however, are composed of multiple distinct experiments. As such, drilldowns on a perturbation gene set contain a Perturbation linkout tab (similar to what one might find when drilling down on a grid entry within the Publication & Reports or HD Mouse Model portals). This linkout tab will provide direct links to each perturbation experiment contributing to the perturbation gene set. As usual, the corner arrow will take you to the Perturbations portal, with the grid pre-populated with only those perturbation experiments contributing to the gene set. See picture below:
Access the HD Explorer:


