HD Explorer
Disease Signatures
GWAS & DNA Repair
Mouse Allelic Series
Data Analysis
|
Integrated network of HD experimental data curated and analyzed from the literature, community ‘omics repositories and newly-released internal CHDI reports. Users can browse and search the network through a HD web application and can download component datasets.
Credits: Data Curation by Rancho BioSciences. Application development by Bridlewood Consulting. |
|
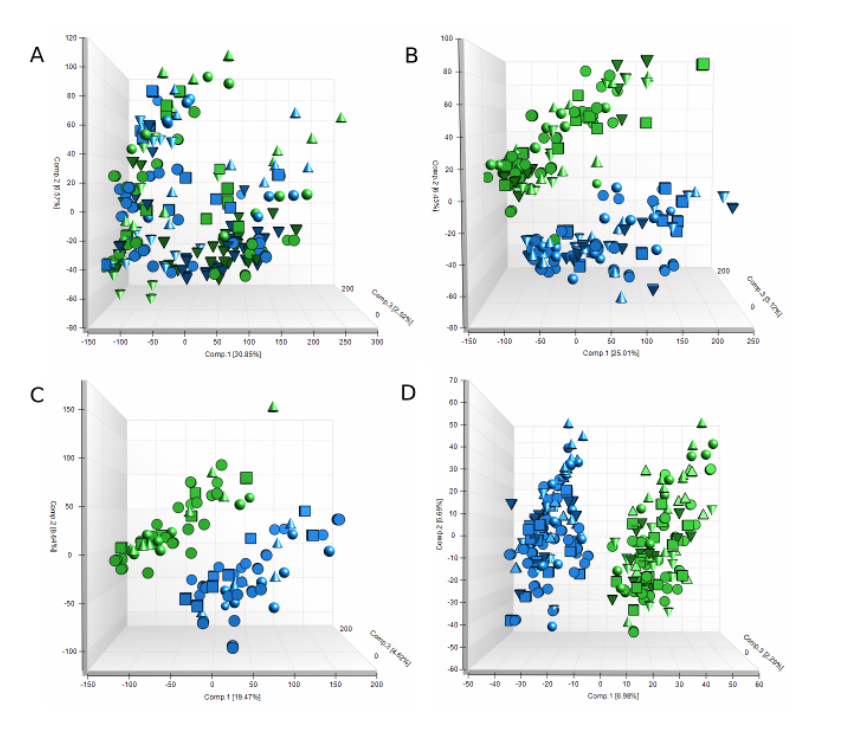
A 266 striatal gene HD disease signature and a 115 striatal protein HD disease signature were developed through analysis of Mouse Allelic Series data and validated in external datasets.
Credits: Data Analysis by Rancho BioSciences. |

| Summary findings from Huntington's disease genome-wide association studies that seek out genes that influence the pathogenesis and expression of Huntington’s disease |
|
Visualization tools and summary findings from Huntington’s disease genetic modifiers analyses.
Credits: GeM MOA Consortium. |

|
Gene reports generated on genes under peaks implicated in early GWAS analyses.
Credits: Data Curation by Rancho BioSciences. |
|
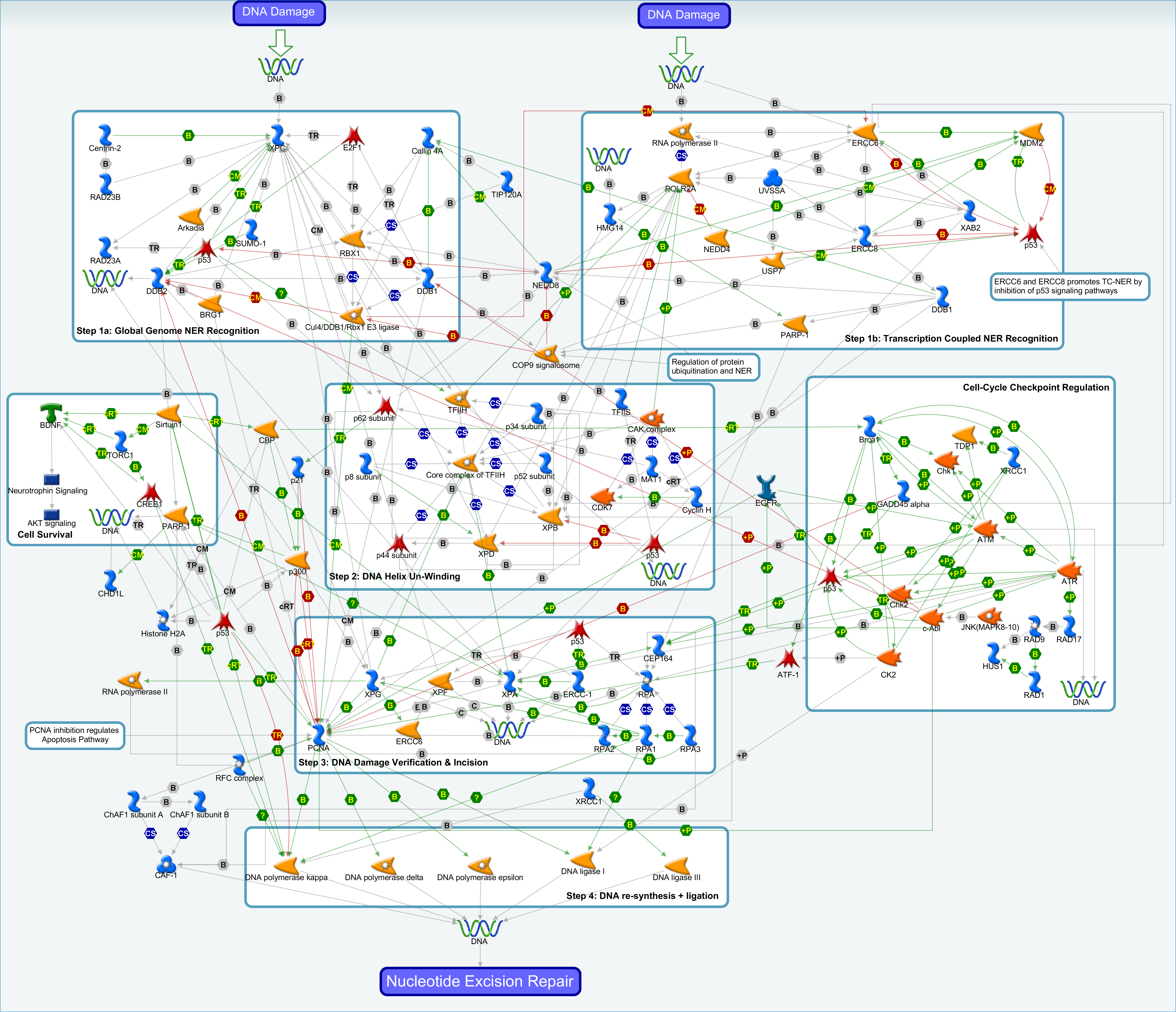
DNA Repair & Handling topic report as well as computable and visual pathways, including DNA Mismatch Repair.
Credits: Clarivate Analytics. |
|
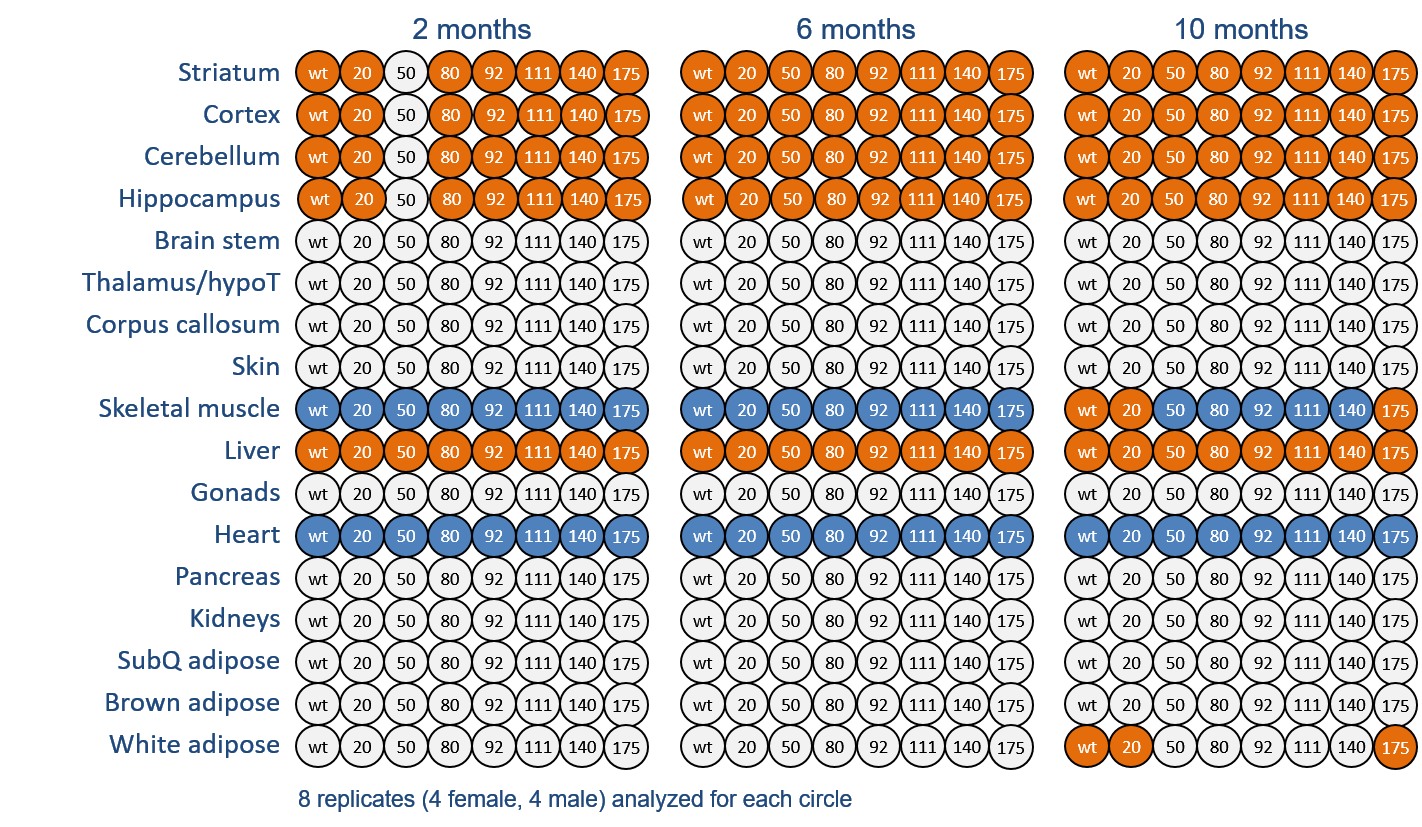
A cross-sectional mouse in-vivo project developed collaboratively between MGH, Psychogenics (PGI) and CHDI. RNAseq, proteomics and behavioural data were generated from both central and peripheral tissue of both WT and an allelic series of HD model mice Q20, Q80, Q92, Q111, Q140, Q175 at 2, 6 and 10 months of age. Data was then subject to differential expression analysis and causal modeling.
Credits: Data Analysis Rancho BioSciences. Causal modeling by AITIA (formerly GNS HealthCare). Mouse Allelic Series project conception and execution: MGH, PsychoGenics (PGI) and CHDI. |
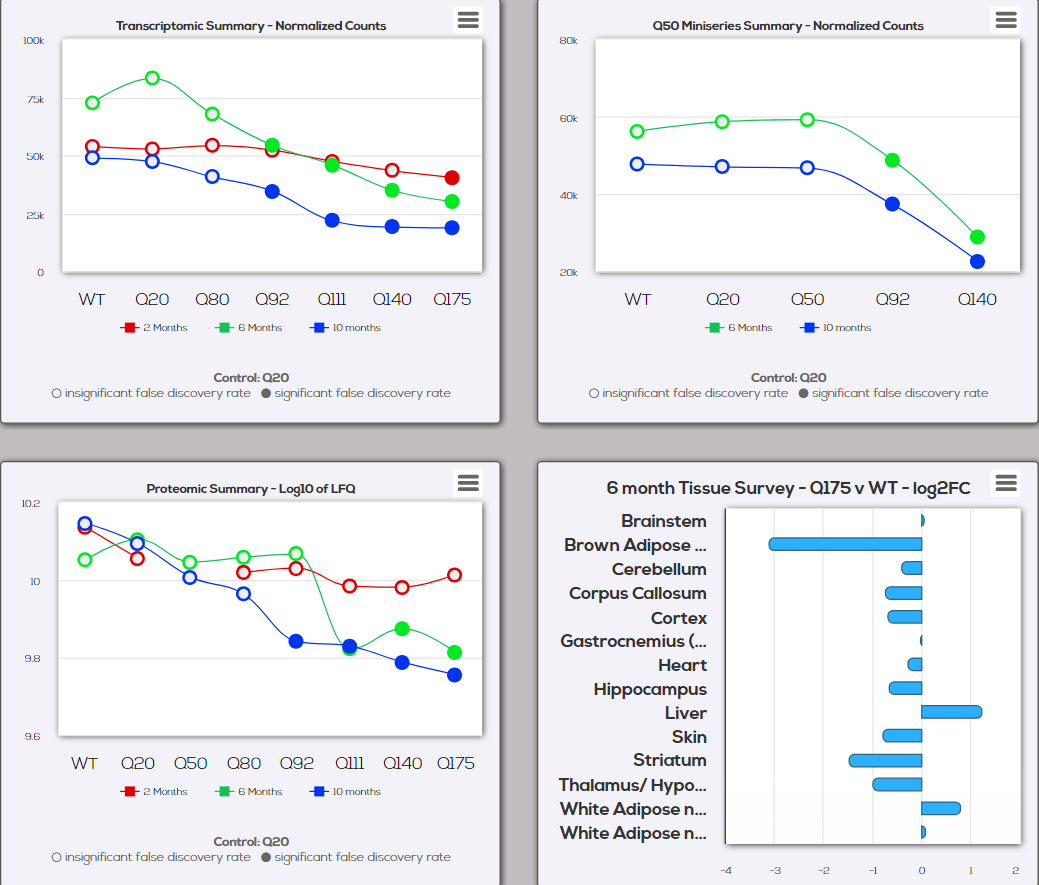
| Visualize tissue-specific mRNA and protein expression in the Mouse Allelic Series at both the aggregate (grouped by genotype) and individual animal level. New in v1.9: CAG length data (from tail or ear) now included for hundreds of individual animals. Credits: Originally developed by David Lipson. Currently maintained by Rancho BioSciences. |
| Visualization tool allowing exploration of results and underlying data of a large scale Weighted Gene Co-expression Network Analysis (WGCNA) of hundreds of samples from intact mouse striatum at 6-month of age as well as from gene set enrichment analysis of transcriptomic signatures of differentially expressed genes from 52 heterozygous HD knockout mice and wildtype controls. Credits: William Yang Lab at UCLA. Application Development: Rancho BioSciences. |
| The Brain-C lab HD knowledge base provides a suite of precision machine learning tools for Huntington’s disease research. Credits: Neri lab (Sorbonne Université and INSERM). |
| Visualization tool providing Astrocyte gene expression profiles across brain regions and HD disease models. Credits: Khakh lab (UCLA). |
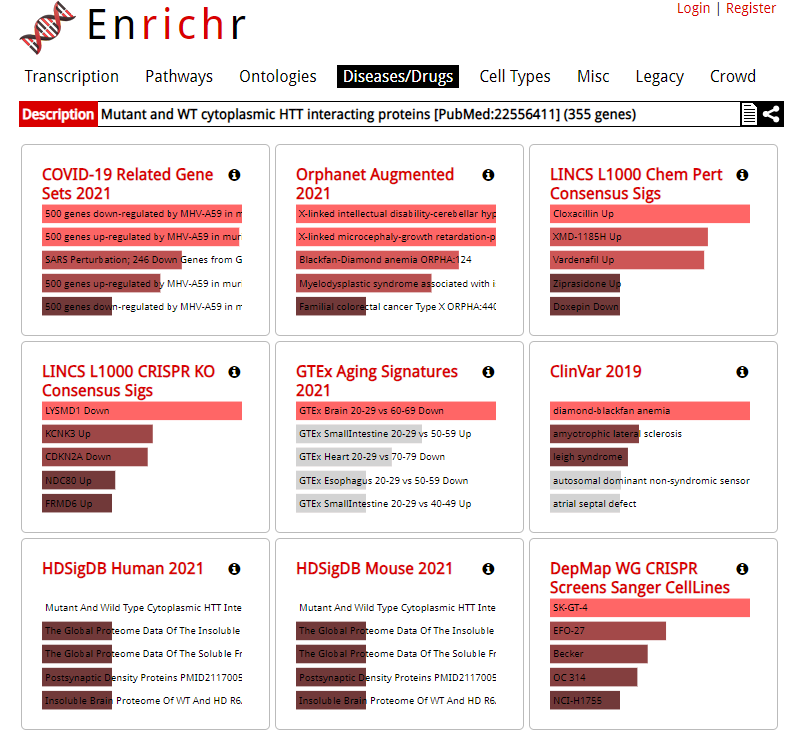
| Gene set enrichment analysis tool operating over a large and diverse collection of gene set libraries including HDSigDB, a gene set library containing HD and HD-related gene sets. Credits: Ma'ayan Lab (Icahn School of Medicine, Mount Sinai). |
| Visualize tissue-specific mRNA and protein expression in the Mouse Allelic Series at both the aggregate (grouped by genotype) and individual animal level. New in v1.9: CAG length data (from tail or ear) now included for hundreds of individual animals. Credits: Originally developed by David Lipson. Currently maintained by Rancho BioSciences. |
| Integrated network of HD experimental data curated and analyzed from the literature, community ‘omics repositories and newly-released internal CHDI reports. | |
| Download component datasets from HD Explorer, including HDSigDB, an HD-relevant gene set enrichment library designed to provide rich functional context for HD-related gene set enrichment analysis. | |
| A 266 striatal gene HD disease signature and a 115 striatal protein HD disease signature were developed through analysis of Mouse Allelic Series data and validated in external datasets. Link to a bioRxiv manuscript describing this work, or download manuscript and supplement files directly. | |
| Summary findings from Huntington's disease genome-wide association studies that seek out genes that influence the pathogenesis and expression of Huntington’s disease | |
| Visualization tools and summary results of a genome-wide association study to identify genetic modifiers of Huntington's disease conducted by the GeM-HD Consortium (PMID: 31398342 and 34180418) | |
| Gene reports generated on genes under peaks implicated in early GWAS analyses. | |
| A topic report (2016) identifying and articulating the genes and pathways involved in DNA damage and repair, with an emphasis of Huntington’s Disease and other trinucleotide disorders. Visual and computational pathway maps are included for DNA Mismatch Repair, Base Excision Repair, Nucleotide Excision Repair and Inter-Strand Crosslink Repair. | |
| Download differential gene and protein expression results from central and peripheral tissues generated within a cross-sectional mouse in-vivo project of both WT and an allelic series of HD model mice with increasingly long CAG repeats (Q20, Q80, Q92, Q111, Q140, Q175) at 2, 6 and 10 months of age. Behavioural data is also available for download. | |
| Gene reports generated on genes under peaks implicated in early GWAS analyses. | |
| Visualize Q-length and age dependent gene and protein expression data from central and peripheral tissue of the cross-sectional Mouse Allelic Series project at both the aggregate (grouped by genotype) and individual animal level. New in v1.9: CAG length data (from tail or ear) now included for hundreds of individual animals. | |
| Visualization tool allowing exploration of results and underlying data of a large scale Weighted Gene Co-expression Network Analysis (WGCNA) of hundreds of samples from intact mouse striatum at 6-month of age as well as from gene set enrichment analysis of transcriptomic signatures of differentially expressed genes from 52 heterozygous HD knockout mice and wildtype controls. | |
| A suite of precision machine learning tools for Huntington’s disease research. | |
| Visualize Astrocyte gene expression profiles across brain regions and HD disease models | |
| Gene set enrichment analysis tool operating over a large and diverse collection of gene set libraries including HDSigDB, a gene set library containing HD and HD-related gene sets. |