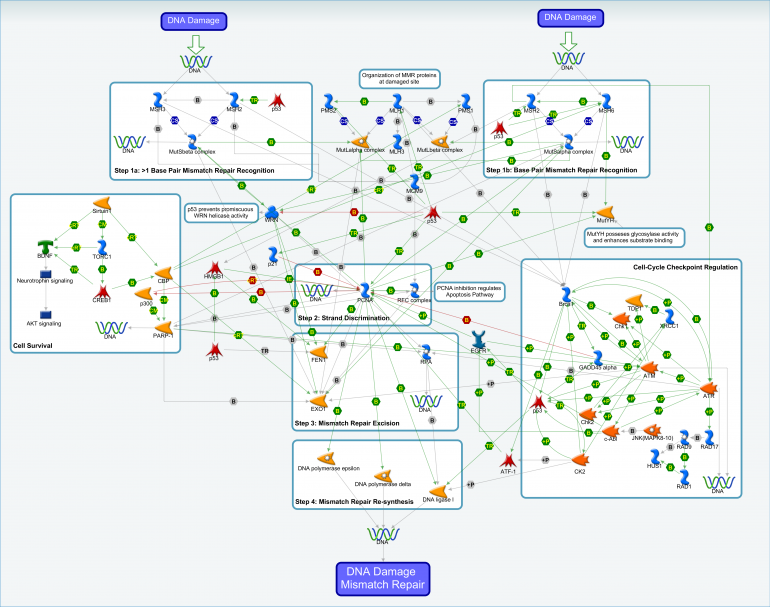
A genome wide association study has implicated a number of DNA damage response pathway members as candidate modifiers of age at motor onset for Huntington’s Disease gene expansion carriers (GeM HD Consortium 2015). In addition, preclinical data in mouse and fly models has shown loss of function mutations in DNA damage response genes can alter somatic expansion rates, mutant huntingtin protein aggregate distribution and animal movement phenotypes (Lu et al, 2014); mutant huntingtin itself also appears to participate in a DNA damage response (e.g. Enokido et al 2010). Extensive gene expression profiling in the mouse allelic series has described CAG length related modules enriched for DNA damage response pathways and p53 signaling (Langfelder et al 2016). The candidate genes described in all of these studies include proteins which mediate a DNA damage response via interstrand cross link repair (ICL), mismatch repair (MMR), base excision repair (BER), or nucleotide excision repair (NER), and there is evidence many of these candidates cross talk between these pathways via direct interactions with each other.
DNA damage response pathway members play important roles in the maintenance of both nuclear and mitochondrial DNA integrity in neurons and other cell types. Recognizing the components of the signaling pathways involved in the DNA damage response is critical to interrogating this mechanism in the context of Huntington’s Disease. We have engaged Clarivate Analytics to generate a literature review of the DNA damage response, visual representations of four pathways (ICL, MMR, BER and NER), and gene lists of the participants within those pathways. The reports, maps and gene lists are now available on HDinHD. Additional mechanisms and pathways will be arriving soon to our “Mechanism Deep Dive” page.
- , , , , , , , , , , , , , , , , , May 3, 2010 216:2

- Langfelder, P., Cantle, J.P., Chatzopoulou, D., Wang, N., Gao, F., , Al-Ramahi, Lu, X.H., Ramos, E.M., El-Zein, K., Zhao, Y., Deverasetty, S., Tebbe, A., Schaab, C., Lavery, D.J., Howland, D., Kwak, S., Botas, J., Aaronson, J.S., Rosinski, J., Coppola, G., Horvath, S., and Yang, X.W. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice, Nat Neurosci., 19, 623–633
If you have an active account on HDinHD, access the DNA Damage Response Data:
To apply for an account on HDinHD:


