Project Report
There is growing evidence from genome-wide association studies in people and experimental testing in model systems that DNA damage is not only a consequence of but also is a significant contributor to the development of neurodegenerative diseases such as Huntington’s Disease. DNA repair pathways play an important role in the maintenance of both nuclear and mitochondrial DNA integrity in neurons and other cell types. Understanding the signaling pathways involved in neuronal DNA damage response is critical to interrogating this mechanism. CHDI has engaged Clarivate Analytics to explore the state of the scientific literature (up to 2016) to identify and articulate the genes and pathways involved in DNA Damage Response and Repair with a special emphasis on the effect in Huntington’s Disease and other trinucleotide repeat disorders.
Project Results
Base Excision Repair
Base Excision Repair (BER) is a repair system resulting from damaged bases in DNA that are subsequently removed and repaired. Two complementary pathways exist in BER, the long-patch and short-patch pathways that repair >2 bases and single base repairs respectively. Direct damage is recognized by glycosylase enzymes, which is followed by the excision of the phosphodiester bond by the AP endonuclease (APE1) at the 5′ side of the damaged site leaving a gap. The small gap left in the DNA helix is filled in by the sequential action of DNA polymerase and DNA ligase, using the undamaged strand as a template.
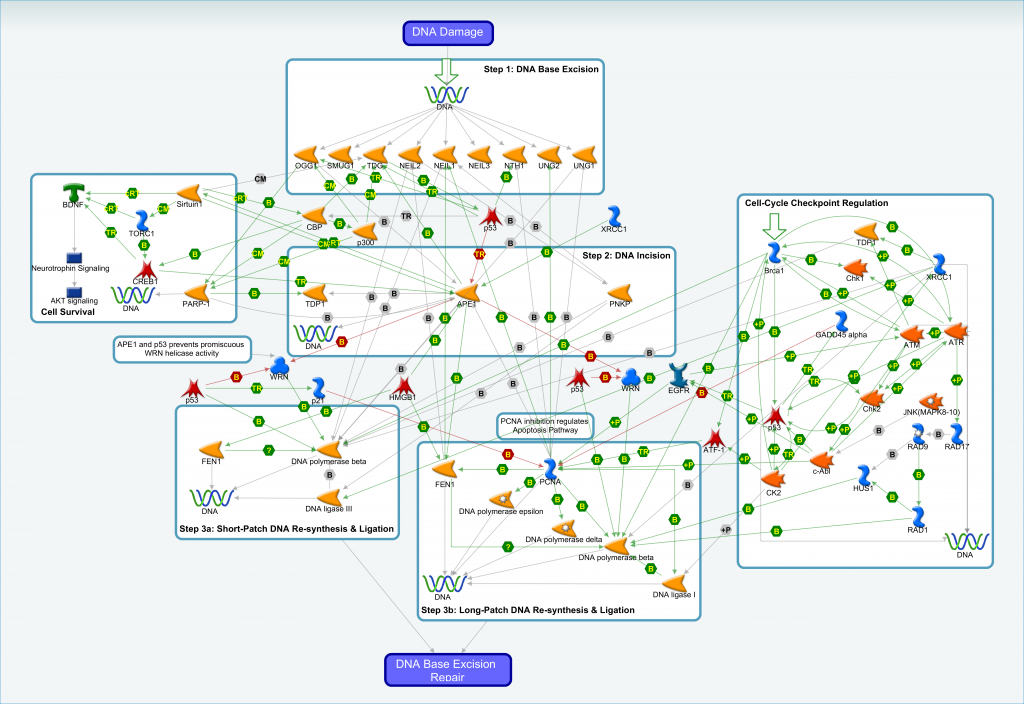
An archive file containing the interaction data for the base excision repair pathway is available. The contents of the archive file include:
- Text and Excel versions of the interaction data
- Zoomable image of the BER pathway shown above
- Reference guide to aid interpretation of visual pathway
Nucleotide Excision Repair
Nucleotide Excision Repair (NER) removes helix-distorting DNA lesions and structures from the genome. The serial steps in NER are similar in organisms from unicellular bacteria to complex mammals, and involve (a) recognition of lesions, adducts or structures that disrupt the DNA double helix, (b) removal of a short oligonucleotide containing the DNA lesion, (c) synthesis of a repair patch copying the opposite undamaged strand, and (d) ligation to restore the DNA to its original form. Two sub-pathways exist in NER: (i) Transcription-coupled NER (TC-NER) is dedicated to the removal of lesions from the template DNA strands of actively transcribed genes, and (ii) Global Genome NER (GG-NER), detects and repairs bulky damages in the entire genome, including in the untranscribed regions and silent chromatin.
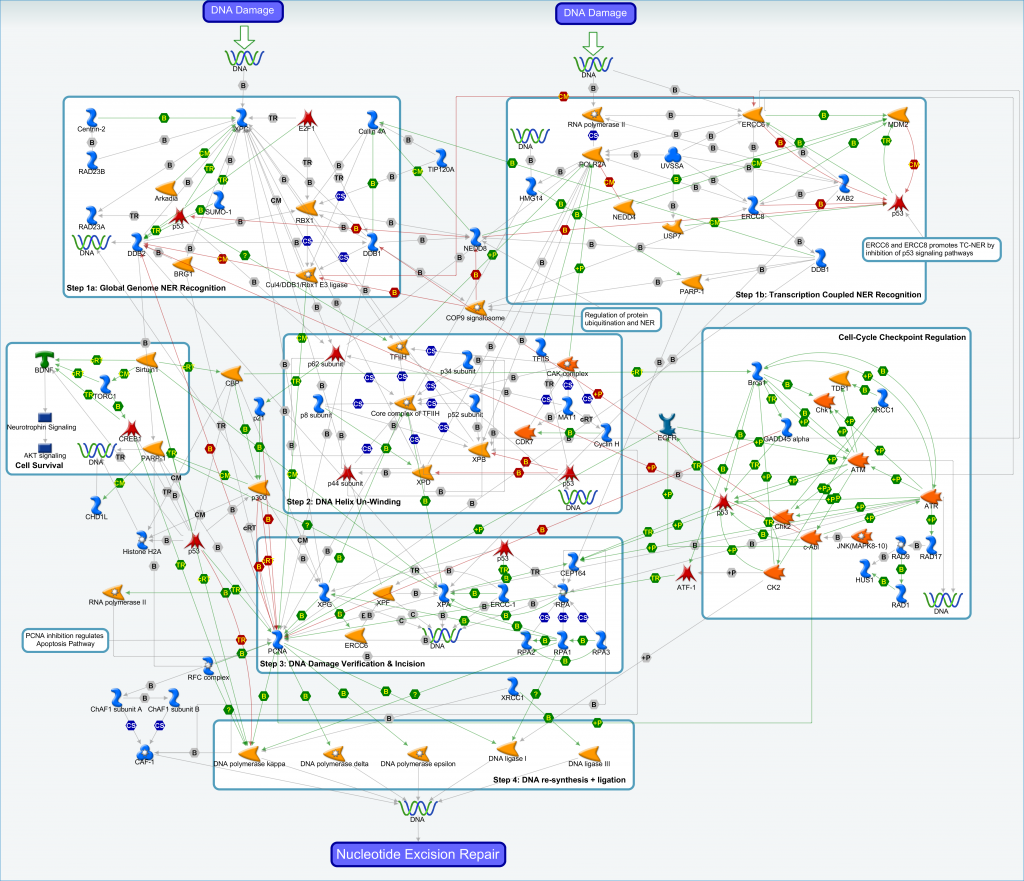
An archive file containing the interaction data for the nucleotide excision repair pathway is available. The contents of the archive file include:
- Text and Excel versions of the interaction data
- Zoomable image of the NER pathway shown above
- Reference guide to aid interpretation of visual pathway
DNA Mismatch Repair
The MMR pathway and the key proteins, MutS and MutL, were originally identified in prokaryotes and are highly conserved in both prokaryotes and eukaryotes. The central steps involved in MMR of DNA damage are a) recognition and initiation of mismatch/damage by MutS and MutL proteins, and b) excision of the mismatch nucleotides by endonucleases, and c) the re-synthesis of the DNA strand by polymerases and ligases.
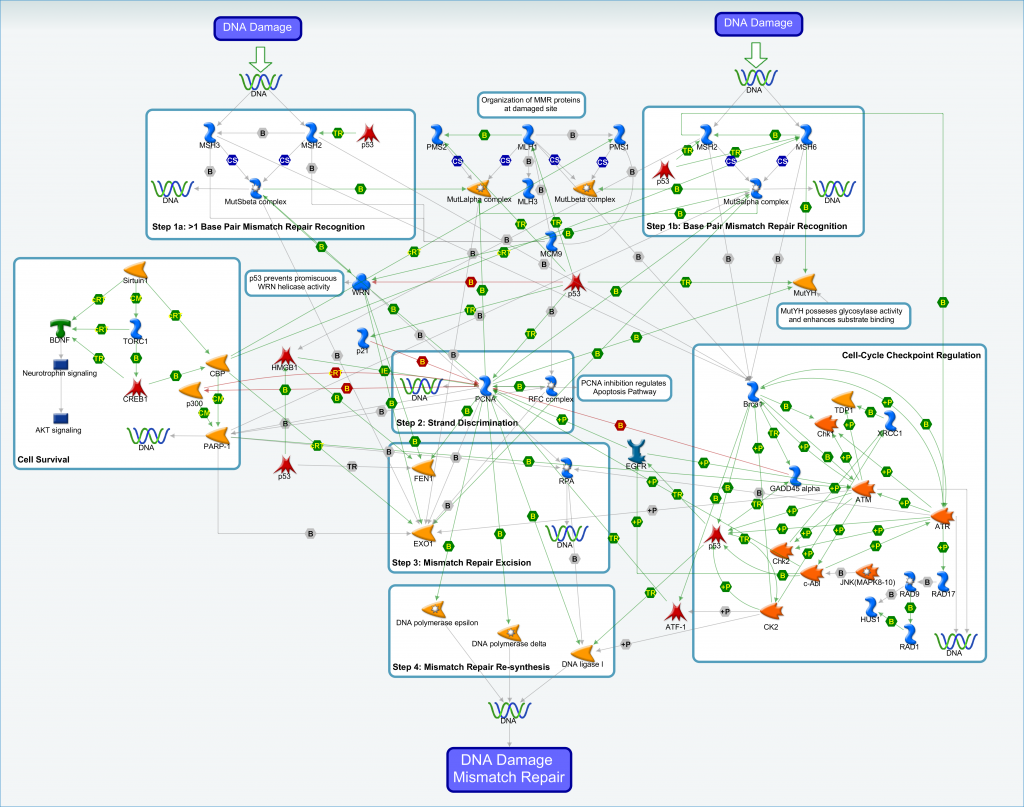
An archive file containing the interaction data for the mismatch repair pathway is available. The contents of the archive file include:
- Text and Excel versions of the interaction data
- Zoomable image of the MMR pathway shown above
- Reference guide to aid interpretation of visual pathway
Inter-Strand Crosslink Repair
Inter-strand DNA crosslinks (ICLs) are covalently linked lesions that form between opposing strands of double-stranded DNA, thereby preventing transcription and replication by inhibition of DNA strand separation. ICLs form in the presence of bifunctional alkylating agents and are extremely cytotoxic, as even a single ICL in the genome can cause severe defects in a variety of vital DNA metabolic processes. The major ICL repair pathway is coupled to DNA replication as it is triggered by replication fork collision with an ICL. When this happens repair is initiated through the excision of the crosslink from one parental strand, releasing one daughter duplex from the ICL forming a double-stranded DNA break that is subsequently repaired. ICL repair is thus a rare instance in which stalled replication forks undergo programmed collapse.
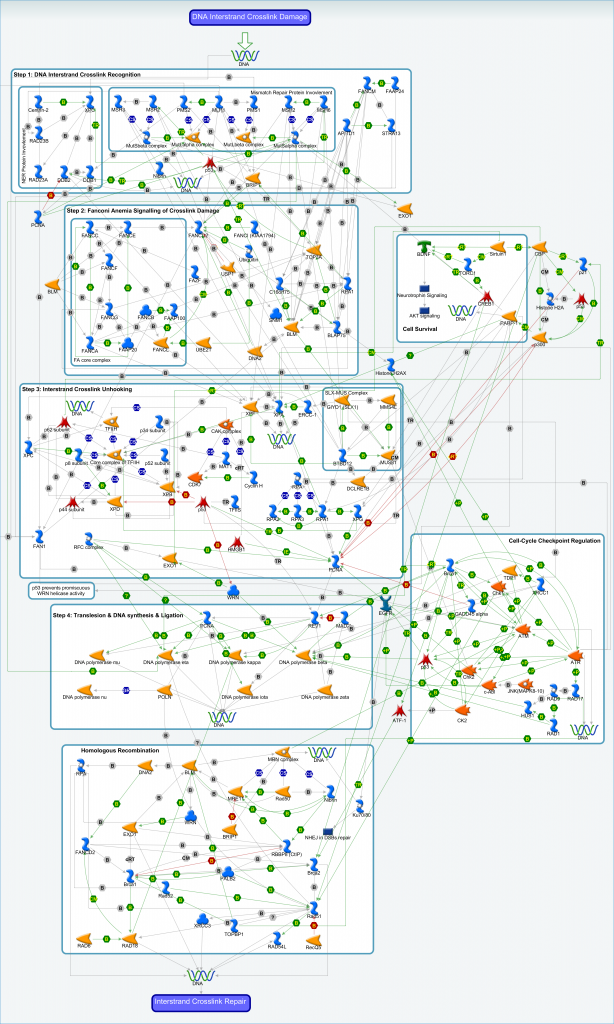
An archive file containing the interaction data for the inter-strand crosslink repair pathway is available. The contents of the archive file include:
- Text and Excel versions of the interaction data
- Zoomable image of the ICL pathway shown above
- Reference guide to aid interpretation of visual pathway
Project Downloads
An archive file containing all interaction data for all DNA Damage Response method types is available. The contents of the archive file include:
- Text and Excel versions of all interaction data
- Zoomable images of all 4 DNA Damage Response pathways
- Reference guide to aid interpretation of visual pathway
- The full report from Clarivate Analytics
- Cytoscape session file containing networks for all method types
- For additional information on Cytoscape network data integration, analysis and visualization and to download the software, please visit cytoscape.org


